paddy.Paddy_Runner module¶
The paddy.Paddy_Runner module contains the
PFARunner class and its associated methods.
Routine listings¶
- PFARunner(object)
Paddy class that runs the paddy field algorithm.
PFARunner.random_step()
PFARunner.run_paddy(self, file_name=None, verbose=None )
Notes¶
The two user defined PFARunner parameters
space and eval_func must be defined such that they are consistent with
the internal operations of paddy. Refer to their specific documentation
detailed in the Parameters section of
PFARunner for more information.
Examples¶
A generic example of using the paddy.Paddy_Runner module
PFARunner and associated methods in a typical
manner. In this case, we are setting up a PFARunner to optimize (x,y)
coordinates for a paraboloid of \(z=-\frac{x^2}{7}-\frac{y^2}{2}+1\).
>>> import paddy
>>> def parabola(input):
... x = input[0][0]
... y = input[1][0]
... return(((x**2)/7)-((y**2)/2)+1)# The maximum is when x and y are 0
...
>>> # now we need to set our parameter space for x and y
>>> x_param = paddy.PaddyParameter(param_range=[-5,5,.2],
... param_type='continuous',
... limits=None, gaussian='scaled',
... normalization = False)
...
>>> y_param = paddy.PaddyParameter(param_range=[-7,3,.2],
... param_type='continuous',
... limits=None, gaussian='scaled',
... normalization = False)
...
>>> # now we make a class with the parameter spaces we defined
>>> class paraboloid_space(object):
... def __init__(self):
... self.xp = x_param
... self.yp = y_param
...
>>> # now we need to initialize a `PFARunner`
>>> example_space = paraboloid_space() #the space parameter
>>> example_runner = paddy.PFARunner(space=example_space,
... eval_func=parabola,
... paddy_type='population',
... rand_seed_number = 20,
... yt = 10,
... Qmax = 5,
... r=.2,
... iterations = 5)
...
>>> example_runner.run_paddy(file_name='paddy_example')
paddy is done!
Lets load the Paddy_Runner and run it some more
>>> import paddy
>>> # we need to define the dependent evaluation function
>>> def parabola(input):
... x = input[0][0]
... y = input[1][0]
... return(((-x**2)/7)-((y**2)/2)+1)# The maximum is when x and y are 0
...
>>> recovered_example = paddy.paddy_recover('paddy_example')
>>> recovered_example.recover_run()
recovered paddy run was already completed, use extend_paddy
>>> # woops! Wrong method, lets use `extend_paddy`!
>>> recovered_example.extend_paddy(5)
paddy is done!
Lets graph the results using
paddy_plot_and_print()
>>> example_runner.paddy_plot_and_print(('scatter',
... 'average_gen',
... 'average_population'))
...
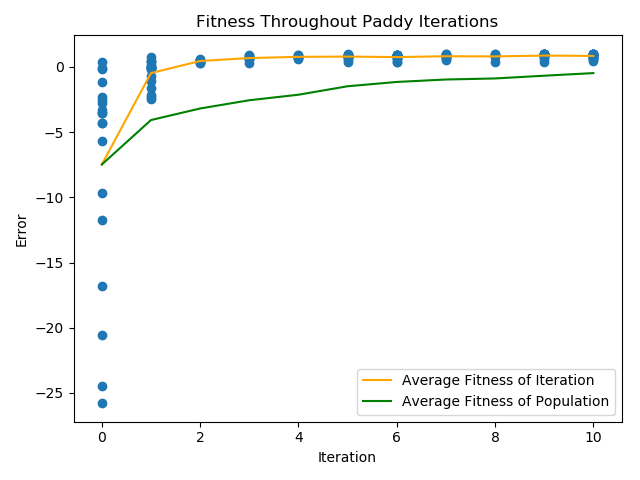
Looks good! But what parameters do the top seed have?
>>> example_runner.get_top_seed()
['seed_116']
>>> example_runner.seed_fitness[116]
0.9961291072913099
>>> example_runner.seed_params[]
array([[-0.29531996, -0.02205951],
[-0.25822639, 0.01529183]])
-
class
paddy.Paddy_Runner.PFARunner(space, eval_func, rand_seed_number, yt, Qmax, paddy_type, r, iterations, error_handling=True)[source]¶ Class that runs PFA.
PFARunner handles the user defined parameters, and exicutes the paddy field algorithm for a defined amount of iterations.
- Parameters
- spaceclass
A user defined class that is comprised of PaddyParameter instances for attributes.
- eval_funcfunction, or method
A function or method that returns a value to be maximized. The function must accept an array-type input where shape = (parameters,2) and reads the zeroth index of every two index lenght slice as a the parameter value where the first index position is the gaussian value which should be ignored.
- paddy_type: string
The string ‘population’ or ‘generational’ to specify which method is used during the selection step.
- rand_seedinteger
A numeric that defines the number of random seeds generated.
- ytinteger
A numeric that defines the threshold operator for the evaluation step.
- Qmaxinteger
A numeric that defines the maximum number of seeds posible that a plant can produce.
- iterationsinteger
A numeric that defines the amount of iterations that paddy will run.
- error_handlingbool, optional (default: True)
A boolean that allows the user to bypass built in error handlings.
- Raises
- PaddyRunnerError
If parameter value(s) used to initialize an instance of PFARunner would raise an exception when calling methods of PFARunner.
- Attributes
- yt_primeinteger or float
A numeric that is defined by the origional yt parameter and is called to reset yt if yt is modified otherwise while running paddy.
- seed_counterinteger
A numeric that tracks the number of seeds generated.
- paddy_counterinteger
A numeric that tracks the number of iteration run.
- seed_paramslist
A list of seed parameters generated while running paddy.
- seed_fitness: list
A list of seed fitness values.
- generation_datadictionary
A dictionary that documents the seed indexes per paddy iteration.
- generation_fitnessdictionary
A dictionary of iterations and the fitness values of their respective seeds.
- top_valuesdictionary
A dictionary of the top seed per iteration, and their respective fitness value and parameters.
- sarray-like, shape = (yt, 2)
A numpy array containing seed indexes and the number of unpolinated seeds.
- Unarray-like, shape = (yt, 2)
A numpy array containing seed indexes and neighbor count values.
- Sarray-like, shape = (yt, 2)
A numpy array containing seed indexes and the number of polinated seeds.
- recoverbool (default
A boolean that determines if the instance of PFARunner has been recovered.
Methods
run_paddy(file_name=None, verbose=None)
Executes the paddy field algorithm. PFARunner methods utilized by run_paddy are described in the respective note section per method.
paddy_plot_and_print(verbose=None)
Used to print, and or graph, results.
save_paddy(new_file_name=None)
Saves the given instance of PFARunner.
recover_run(new_verbose=None)
Resumes the paddy field algorithm if otherwise interupted or recovered.
extend_paddy(new_iterations, new_file_name=None, new_verbose=None)
Extends the iterations of a previously completed instance of PFARunner.
get_top_seed()
Provides the seed of greatest fitness in population.
get_generation_top_seed(counter=None, verbose=False)
Provides the seed of greatest fitness in a generation.
-
extend_paddy(self, new_iterations, new_file_name=None, new_verbose=None)[source]¶ Extend the iterations of a completed paddy runner.
This method runs a PFARunner instance that has completed the amount of iterations set as its attribute.
- Parameters
- new_iterationsinteger
Integer to extend the iterations of the PFARunner by.
- new_file_namestring, optional (default
A string that is used for the file handle. The same string is also used for generating the file handle of the backup pickle with the trailing ‘_backup.pickle’ added, and replaces the former file name used.
- new_verbosestring or sequence of strings, optional (default
See verbose parameter description for run_paddy.
- Raises
- PaddyRunnerError
If parameter value(s) used to initialize an instance of PFARunner would raise an exception when calling methods of PFARunner.
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
recover_run()PFARunner method that runs a recovered pickle run.
Notes
This method can be used to extend the iterations of a PFARunner while changing the file_name and verbose atributes as well in one line rather than manualy for each and calling
run_paddy().
-
get_generation_top_seed(self, counter=None, verbose=False)[source]¶ Return the max value of fitness in the generation.
This method returns the top seed number of a given generation of a paddy run when called internally, though a user can call the method directly to print to terminal. If called directly, a counter parameter value can be passed to specify which generation is acessed, where the default generation analyzed is the last evaluated.
- Parameters
- counterinteger, (default
An integer that specifies which generation is analyzed.
- verbosebool, (default
- Returns
- temp_nameslist of strings
A list of seed number string(s) in the form ‘seed_#’
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
Notes
When running paddy via
run_paddy(), theverboseparameter will callget_generation_top_seed()if the string command ‘top_gen’ is passed.In the case that multiple seeds are tied for having the greatest fitness value, they are appended to temp_names.
-
get_top_seed(self)[source]¶ Return the seed with the greatest fitness in the paddy runner.
This method returns the top seed number when called internally, though a user can simply call the method directly to print to terminal.
- Returns
- temp_nameslist of strings
A list of seed number string(s) in the form ‘seed_#’
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
Notes
When running paddy via
run_paddy(), theverboseparameter will callget_top_seed()if the string command ‘pop’ is passed.In the case that multiple seeds are tied for having the greatest fitness value, they are appended to temp_names.
-
neighbor_counter(self)[source]¶ Generate neighbor counts and polinated seeds.
Calculates the amount of neighbors per seed, and then uses the neighbor counts to calculate the polination factor per seed. Polinated seed values are defined as the rounded integer resulting from the product of the unpolinated seed term and polination term.
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
paddy.Paddy_Parameter.PaddyParameter.get_ecludian_values()Function that retrieves values for calculating ecludian distance of parameters.
Notes
Polinated seeds are calculated as the product of unpolinated seed numbers (s) and the polination factor (U). The polination factor is varient and defined by the number of ‘neighbors’ within the population of seeds being evaluated. Neighbors (u) are defined as seeds falling within the radius (r) of a multi-dimensional euclidean space as such:
\[u(x_{j},x_{k})=\left \| x_{j}-x_{k} \right \|-r<0\]After calculating the number of neighbors per seed in the evaluated population, a polination factor is then calculated and assigned as a normalized value falling between zero and one via the expression:
\[U=e^{[{u\over u_{max}}-1]}\]If the radius parameter provided by the user fails to generate any neighbors for the evaluated seeds, an initial quantile cutoff of 0.75 is applied to an internal list of euclidean distances to generate a new radius term for neighbor counting. This corrective procedure itterates untill the evaluated population contains seeds with neighbors, with the quantile cutoff decreasing by 0.05 untill a cutoff of 0.05 is applied. In the case that this fails, the number of neighbors per seed is simply assigned as one, effectively dropping the polination factor in the current iteration.
-
new_propogation(self)[source]¶ Generate new seeds and evaluate.
Assignes parameters to new seeds, and evaluates them to generate respective fitness values.
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
paddy.Paddy_Parameter.PaddyParameter.new_seed_init()PaddyParameter method that generates new seed parameters.
-
paddy_plot_and_print(self, verbose=None, figure_name=None)[source]¶ Plot and print results.
This method is used to generate results as either printed or plotted results. The type of output is dictated by passed strings via the verbose parameter.
- Parameters
- verbosestring or sequence of string, optional (default
String comand(s) that handle printing of values generated after completion of a paddy run. Details regarding valid strings are discussed in the notes section below.
- figure_namestring, (default
A string that is passed to writer.preformance_plotter that is used to save generated plots.
See also
Notes
paddy_plot_and_print coordinates with writer.preformance_plotter to generate desired representations of results via string arguments initially introduced via verbose. Valid strings that will generate output without calling writer.preformance_plotter are:
- ‘generation’
Simply prints the PFARunner attribute
generation_data.
- ‘final_results’
Prints results via clean_parameter_print with the PFARunner attribute
top_valuesas input.
- ‘pop_fitness’
Prints the list returned by get_top_fitness using the PFARunner attribute
top_valuesas input.
- ‘gen_fitness’
Prints the PFARunner attribute
generation_fitness.
Valid comands are passed to writer.preformance_plotter as described in detail in the notes section for
paddy.writer.preformance_plotter().
-
random_step(self)[source]¶ Generate random seeds.
Excecutes the initialization step of the paddy field algorithm via random propogation of PaddyParameter attributes in provided space.
See also
paddy.utils.random_propogation()function that generates random seeds and an updated seed counter.
run_paddy()PFARunner method that excecutes the paddy field algorithm.
-
recover_run(self, new_verbose=None)[source]¶ Run paddy after recovering a pickled run.
This method runs a recovered depickled PFARunner instance, by resuming where the algorithm was functioning last.
- Parameters
- new_verbosestring or sequence of strings, optional (default
See verbose parameter description for run_paddy.
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
paddy.utils.paddy_recover()utils function that depickles a given pickled PFARunner instance.
Notes
If a recovered PFARunner is unable to excicute
recover_run(), it may be needed to manualy decrease the paddy_counter attribute.
-
run_paddy(self, file_name=None, verbose=None)[source]¶ Excecute paddy field algorithm :func:’run_paddy’.
This method is used to run the paddy field algorithm. A thorough explination of how the method works can be found in the notes section of the
paddy.Paddy_Runnermodule.- Parameters
- file_namestring or nonetype, optional (default
A user defined string that is used as the file handle for PFARunner instance pickling.
- verbosestring or sequence of string, optional (default
String comand(s) that handle printing of values generated during paddy runs. Valid strings include:
('status','top_gen','pop','all').
See also
random_step()Method that excecutes the random initiation step.
sowing_function()Method that calculates unpolinated seed values.
neighbor_counter()Method that calculates neighbor terms and the resulting polination term.
new_propogation()Method that generates new seeds and evaluates them.
paddy.Paddy_Parameter.PaddyParameterClass that handles calculations for specific parameters being optimized by paddy and communicates with PFARunner methods.
Notes
For specifics regarding the dependant methods and
PaddyParameter, refer to their respective documentation.Examples
Setting up a paddy run using default Polynomial parameter space generator function to optimize parameters for trigonometric polynomial interpolation of the Gramacy & Lee function.
>>> import paddy >>> from paddy.Default_Numerics import *
The default polynomial class is used to set up parameter space:
>>> polly_space = Polynomial(length=15, scope=1, ... gaussian_type='scaled', ... normalization=False)
A class instance with an evaluation method for optimization:
>>> class RunFunc(object): ... def __init__(self): ... self.xs , self.ys = gramacy_lee() ... def eval(self,values): ... score = eval_gl(seed=values,x=self.xs,y=self.ys) ... return score >>> rf = RunFunc()
A PFARunner instance is generated and run using run_paddy:
>>> runner = paddy.PFARunner(space=polly_space, ... eval_func=rf.eval, ... rand_seed_number = 50, ... yt = 10, paddy_type='population', ... Qmax=50, r=0.75, iterations=10)
PFARunner parameters can be changed before and after runs:
>>> runner.Qmax = 10 >>> runner.Qmax 10
The PFARunner instance can then run the paddy field algorithm:
>>> runner.run_paddy() paddy is done!
-
save_paddy(self, new_file_name=None)[source]¶ Save
PFARunneras a pickled file.This method uses the python pickle serialization module to store class iterances of PFARunner to be recovered as needed by the user.
- Parameters
- new_file_namestring, (default
A string that is used for the file handle. The same string is also used for generating the file handle of the backup pickle with the trailing ‘_backup.pickle’ added, and replaces the former file name used.
See also
paddy.utils.paddy_recover()utils function that depickles a given pickled PFARunner instance.
Notes
The backup pickle is saved every other iteration of paddy after the primary pickle handle.
-
sowing_function(self)[source]¶ Generate unpolinated seeds.
Applies the threshold operator yt on the seeds to be considered, defined by the paddy type, and calculates the number of unpolinated seeds for the subsequent steps in the paddy field algorithm.
See also
run_paddyPFARunner method that excecutes the paddy field algorithm.
Notes
The user defined threshold operator yt is used to select seeds to be subsequently considered for the remaining steps of the current paddy itteration, where yt is an integer that represents the nth fittest seed. However, if yt is greater than the number of seeds being considered, yt is redefined as:
\[y_{t}\simeq 0.75 \ast y_{considered}\]New seeds (s) are then generated for evaluated seeds of fitness greater than yt using min-max feature scalling to the form of:
\[s=Q_{max^{\ast}}[{y-y_{t}\over y_{max}-y_{t}}]\]
-
class
paddy.Paddy_Runner.PFARunner(space, eval_func, rand_seed_number, yt, Qmax, paddy_type, r, iterations, error_handling=True)[source] Bases:
objectClass that runs PFA.
PFARunner handles the user defined parameters, and exicutes the paddy field algorithm for a defined amount of iterations.
- Parameters
- spaceclass
A user defined class that is comprised of PaddyParameter instances for attributes.
- eval_funcfunction, or method
A function or method that returns a value to be maximized. The function must accept an array-type input where shape = (parameters,2) and reads the zeroth index of every two index lenght slice as a the parameter value where the first index position is the gaussian value which should be ignored.
- paddy_type: string
The string ‘population’ or ‘generational’ to specify which method is used during the selection step.
- rand_seedinteger
A numeric that defines the number of random seeds generated.
- ytinteger
A numeric that defines the threshold operator for the evaluation step.
- Qmaxinteger
A numeric that defines the maximum number of seeds posible that a plant can produce.
- iterationsinteger
A numeric that defines the amount of iterations that paddy will run.
- error_handlingbool, optional (default: True)
A boolean that allows the user to bypass built in error handlings.
- Raises
- PaddyRunnerError
If parameter value(s) used to initialize an instance of PFARunner would raise an exception when calling methods of PFARunner.
- Attributes
- yt_primeinteger or float
A numeric that is defined by the origional yt parameter and is called to reset yt if yt is modified otherwise while running paddy.
- seed_counterinteger
A numeric that tracks the number of seeds generated.
- paddy_counterinteger
A numeric that tracks the number of iteration run.
- seed_paramslist
A list of seed parameters generated while running paddy.
- seed_fitness: list
A list of seed fitness values.
- generation_datadictionary
A dictionary that documents the seed indexes per paddy iteration.
- generation_fitnessdictionary
A dictionary of iterations and the fitness values of their respective seeds.
- top_valuesdictionary
A dictionary of the top seed per iteration, and their respective fitness value and parameters.
- sarray-like, shape = (yt, 2)
A numpy array containing seed indexes and the number of unpolinated seeds.
- Unarray-like, shape = (yt, 2)
A numpy array containing seed indexes and neighbor count values.
- Sarray-like, shape = (yt, 2)
A numpy array containing seed indexes and the number of polinated seeds.
- recoverbool (default
A boolean that determines if the instance of PFARunner has been recovered.
Methods
run_paddy(file_name=None, verbose=None)
Executes the paddy field algorithm. PFARunner methods utilized by run_paddy are described in the respective note section per method.
paddy_plot_and_print(verbose=None)
Used to print, and or graph, results.
save_paddy(new_file_name=None)
Saves the given instance of PFARunner.
recover_run(new_verbose=None)
Resumes the paddy field algorithm if otherwise interupted or recovered.
extend_paddy(new_iterations, new_file_name=None, new_verbose=None)
Extends the iterations of a previously completed instance of PFARunner.
get_top_seed()
Provides the seed of greatest fitness in population.
get_generation_top_seed(counter=None, verbose=False)
Provides the seed of greatest fitness in a generation.
-
__init__(self, space, eval_func, rand_seed_number, yt, Qmax, paddy_type, r, iterations, error_handling=True)[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
__weakref__¶ list of weak references to the object (if defined)
-
extend_paddy(self, new_iterations, new_file_name=None, new_verbose=None)[source] Extend the iterations of a completed paddy runner.
This method runs a PFARunner instance that has completed the amount of iterations set as its attribute.
- Parameters
- new_iterationsinteger
Integer to extend the iterations of the PFARunner by.
- new_file_namestring, optional (default
A string that is used for the file handle. The same string is also used for generating the file handle of the backup pickle with the trailing ‘_backup.pickle’ added, and replaces the former file name used.
- new_verbosestring or sequence of strings, optional (default
See verbose parameter description for run_paddy.
- Raises
- PaddyRunnerError
If parameter value(s) used to initialize an instance of PFARunner would raise an exception when calling methods of PFARunner.
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
recover_run()PFARunner method that runs a recovered pickle run.
Notes
This method can be used to extend the iterations of a PFARunner while changing the file_name and verbose atributes as well in one line rather than manualy for each and calling
run_paddy().
-
get_generation_top_seed(self, counter=None, verbose=False)[source] Return the max value of fitness in the generation.
This method returns the top seed number of a given generation of a paddy run when called internally, though a user can call the method directly to print to terminal. If called directly, a counter parameter value can be passed to specify which generation is acessed, where the default generation analyzed is the last evaluated.
- Parameters
- counterinteger, (default
An integer that specifies which generation is analyzed.
- verbosebool, (default
- Returns
- temp_nameslist of strings
A list of seed number string(s) in the form ‘seed_#’
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
Notes
When running paddy via
run_paddy(), theverboseparameter will callget_generation_top_seed()if the string command ‘top_gen’ is passed.In the case that multiple seeds are tied for having the greatest fitness value, they are appended to temp_names.
-
get_top_seed(self)[source] Return the seed with the greatest fitness in the paddy runner.
This method returns the top seed number when called internally, though a user can simply call the method directly to print to terminal.
- Returns
- temp_nameslist of strings
A list of seed number string(s) in the form ‘seed_#’
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
Notes
When running paddy via
run_paddy(), theverboseparameter will callget_top_seed()if the string command ‘pop’ is passed.In the case that multiple seeds are tied for having the greatest fitness value, they are appended to temp_names.
-
neighbor_counter(self)[source] Generate neighbor counts and polinated seeds.
Calculates the amount of neighbors per seed, and then uses the neighbor counts to calculate the polination factor per seed. Polinated seed values are defined as the rounded integer resulting from the product of the unpolinated seed term and polination term.
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
paddy.Paddy_Parameter.PaddyParameter.get_ecludian_values()Function that retrieves values for calculating ecludian distance of parameters.
Notes
Polinated seeds are calculated as the product of unpolinated seed numbers (s) and the polination factor (U). The polination factor is varient and defined by the number of ‘neighbors’ within the population of seeds being evaluated. Neighbors (u) are defined as seeds falling within the radius (r) of a multi-dimensional euclidean space as such:
\[u(x_{j},x_{k})=\left \| x_{j}-x_{k} \right \|-r<0\]After calculating the number of neighbors per seed in the evaluated population, a polination factor is then calculated and assigned as a normalized value falling between zero and one via the expression:
\[U=e^{[{u\over u_{max}}-1]}\]If the radius parameter provided by the user fails to generate any neighbors for the evaluated seeds, an initial quantile cutoff of 0.75 is applied to an internal list of euclidean distances to generate a new radius term for neighbor counting. This corrective procedure itterates untill the evaluated population contains seeds with neighbors, with the quantile cutoff decreasing by 0.05 untill a cutoff of 0.05 is applied. In the case that this fails, the number of neighbors per seed is simply assigned as one, effectively dropping the polination factor in the current iteration.
-
new_propogation(self)[source] Generate new seeds and evaluate.
Assignes parameters to new seeds, and evaluates them to generate respective fitness values.
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
paddy.Paddy_Parameter.PaddyParameter.new_seed_init()PaddyParameter method that generates new seed parameters.
-
paddy_plot_and_print(self, verbose=None, figure_name=None)[source] Plot and print results.
This method is used to generate results as either printed or plotted results. The type of output is dictated by passed strings via the verbose parameter.
- Parameters
- verbosestring or sequence of string, optional (default
String comand(s) that handle printing of values generated after completion of a paddy run. Details regarding valid strings are discussed in the notes section below.
- figure_namestring, (default
A string that is passed to writer.preformance_plotter that is used to save generated plots.
See also
Notes
paddy_plot_and_print coordinates with writer.preformance_plotter to generate desired representations of results via string arguments initially introduced via verbose. Valid strings that will generate output without calling writer.preformance_plotter are:
- ‘generation’
Simply prints the PFARunner attribute
generation_data.
- ‘final_results’
Prints results via clean_parameter_print with the PFARunner attribute
top_valuesas input.
- ‘pop_fitness’
Prints the list returned by get_top_fitness using the PFARunner attribute
top_valuesas input.
- ‘gen_fitness’
Prints the PFARunner attribute
generation_fitness.
Valid comands are passed to writer.preformance_plotter as described in detail in the notes section for
paddy.writer.preformance_plotter().
-
random_step(self)[source] Generate random seeds.
Excecutes the initialization step of the paddy field algorithm via random propogation of PaddyParameter attributes in provided space.
See also
paddy.utils.random_propogation()function that generates random seeds and an updated seed counter.
run_paddy()PFARunner method that excecutes the paddy field algorithm.
-
recover_run(self, new_verbose=None)[source] Run paddy after recovering a pickled run.
This method runs a recovered depickled PFARunner instance, by resuming where the algorithm was functioning last.
- Parameters
- new_verbosestring or sequence of strings, optional (default
See verbose parameter description for run_paddy.
See also
run_paddy()PFARunner method that excecutes the paddy field algorithm.
paddy.utils.paddy_recover()utils function that depickles a given pickled PFARunner instance.
Notes
If a recovered PFARunner is unable to excicute
recover_run(), it may be needed to manualy decrease the paddy_counter attribute.
-
run_paddy(self, file_name=None, verbose=None)[source] Excecute paddy field algorithm :func:’run_paddy’.
This method is used to run the paddy field algorithm. A thorough explination of how the method works can be found in the notes section of the
paddy.Paddy_Runnermodule.- Parameters
- file_namestring or nonetype, optional (default
A user defined string that is used as the file handle for PFARunner instance pickling.
- verbosestring or sequence of string, optional (default
String comand(s) that handle printing of values generated during paddy runs. Valid strings include:
('status','top_gen','pop','all').
See also
random_step()Method that excecutes the random initiation step.
sowing_function()Method that calculates unpolinated seed values.
neighbor_counter()Method that calculates neighbor terms and the resulting polination term.
new_propogation()Method that generates new seeds and evaluates them.
paddy.Paddy_Parameter.PaddyParameterClass that handles calculations for specific parameters being optimized by paddy and communicates with PFARunner methods.
Notes
For specifics regarding the dependant methods and
PaddyParameter, refer to their respective documentation.Examples
Setting up a paddy run using default Polynomial parameter space generator function to optimize parameters for trigonometric polynomial interpolation of the Gramacy & Lee function.
>>> import paddy >>> from paddy.Default_Numerics import *
The default polynomial class is used to set up parameter space:
>>> polly_space = Polynomial(length=15, scope=1, ... gaussian_type='scaled', ... normalization=False)
A class instance with an evaluation method for optimization:
>>> class RunFunc(object): ... def __init__(self): ... self.xs , self.ys = gramacy_lee() ... def eval(self,values): ... score = eval_gl(seed=values,x=self.xs,y=self.ys) ... return score >>> rf = RunFunc()
A PFARunner instance is generated and run using run_paddy:
>>> runner = paddy.PFARunner(space=polly_space, ... eval_func=rf.eval, ... rand_seed_number = 50, ... yt = 10, paddy_type='population', ... Qmax=50, r=0.75, iterations=10)
PFARunner parameters can be changed before and after runs:
>>> runner.Qmax = 10 >>> runner.Qmax 10
The PFARunner instance can then run the paddy field algorithm:
>>> runner.run_paddy() paddy is done!
-
save_paddy(self, new_file_name=None)[source] Save
PFARunneras a pickled file.This method uses the python pickle serialization module to store class iterances of PFARunner to be recovered as needed by the user.
- Parameters
- new_file_namestring, (default
A string that is used for the file handle. The same string is also used for generating the file handle of the backup pickle with the trailing ‘_backup.pickle’ added, and replaces the former file name used.
See also
paddy.utils.paddy_recover()utils function that depickles a given pickled PFARunner instance.
Notes
The backup pickle is saved every other iteration of paddy after the primary pickle handle.
-
sowing_function(self)[source] Generate unpolinated seeds.
Applies the threshold operator yt on the seeds to be considered, defined by the paddy type, and calculates the number of unpolinated seeds for the subsequent steps in the paddy field algorithm.
See also
run_paddyPFARunner method that excecutes the paddy field algorithm.
Notes
The user defined threshold operator yt is used to select seeds to be subsequently considered for the remaining steps of the current paddy itteration, where yt is an integer that represents the nth fittest seed. However, if yt is greater than the number of seeds being considered, yt is redefined as:
\[y_{t}\simeq 0.75 \ast y_{considered}\]New seeds (s) are then generated for evaluated seeds of fitness greater than yt using min-max feature scalling to the form of:
\[s=Q_{max^{\ast}}[{y-y_{t}\over y_{max}-y_{t}}]\]